- Get link
- X
- Other Apps
PCR stands for Polymerase Chain Reaction. It is a widely used molecular biology technique that allows scientists to amplify (make many copies of) a specific segment of DNA (deoxyribonucleic acid) or RNA (ribonucleic acid) in a laboratory setting. PCR is essential in various applications, including genetics research, disease diagnosis, forensics, and more. DNA sequence can be analyzed, sequenced, or used for various applications. PCR has revolutionized molecular biology and has found applications in fields such as genetic testing, DNA fingerprinting, microbial detection, and the study of gene expression. There are also variations of PCR, such as reverse transcription PCR (RT-PCR) used for amplifying RNA, quantitative PCR (qPCR) for quantifying DNA or RNA, and digital PCR (dPCR) for absolute quantification of nucleic acids, among others.
- Steps of PCR
- Denaturation:
- The DNA sample is heated to a high temperature (usually around 94-98°C).
- This causes the double-stranded DNA to separate into two single strands by breaking the hydrogen bonds between complementary base pairs (adenine-thymine and cytosine-guanine).
- Annealing:
- The reaction temperature is lowered to a specific range (usually between 50-65°C).
- During this step, short DNA sequences called primers, which are designed to be complementary to the sequences flanking the target region, bind (anneal) to the single-stranded DNA.
- Primers provide a starting point for DNA polymerases to begin copying the target DNA.
- Extension:
- The temperature is raised slightly (usually around 72°C).
- DNA polymerase enzyme, often Taq polymerase (isolated from a thermophilic bacterium), adds nucleotides to the primers, synthesizing a new DNA strand complementary to the target region.
- This process extends the DNA chain.
- Repeat:
- Steps 1-3 are repeated multiple times (usually 20-40 cycles) in a thermal cycler machine.
- Each cycle roughly doubles the amount of target DNA.
- This exponential
amplification allows researchers to create millions of copies of the specific
DNA segment of interest.
- Applications
- Genetic Testing:
- PCR is used to detect genetic mutations, analyze gene expression, and study DNA variations.
- It is essential in diagnosing genetic disorders and identifying genetic markers associated with diseases.
- Forensic Science:
- DNA profiling using PCR is a vital tool in forensic investigations.
- It helps identify individuals
based on DNA from crime scenes or human remains.
- Infectious Disease Diagnosis:
- PCR can detect the presence of infectious agents like bacteria, viruses, and parasites in clinical samples.
- It's widely used in diagnosing diseases such as HIV, COVID-19, tuberculosis, and hepatitis.
- Microbial Detection:
- PCR is employed to identify and study microorganisms in various environments, including soil, water, and the human body.
- It helps in microbial ecology and epidemiological studies.
- Food Safety:
- PCR can detect foodborne pathogens, ensuring the safety of food products.
- It's also used in verifying the authenticity of food products, such as confirming the presence of specific ingredients in processed foods.
- Variations of PCR
- Reverse Transcription PCR (RT-PCR):
- This variation is used to amplify RNA rather than DNA.
- It's an essential tool for studying gene expression because it allows researchers to convert RNA into complementary DNA (cDNA) and then amplify specific cDNA sequences.
- This technique is commonly
used to measure RNA levels and analyze gene expression patterns.
- Quantitative PCR (qPCR):
- Also known as real-time PCR, qPCR allows for the quantification of the initial amount of DNA or RNA in a sample.
- It's used in gene expression analysis, viral load measurements, and more.
- Real-Time PCR, also known as quantitative PCR (qPCR), is used to measure the amount of DNA or RNA in a sample in real-time during the PCR reaction.
- It allows for the precise
quantification of target molecules and is commonly used for gene expression
analysis, viral load quantification, and DNA quantification in applications
like genotyping.
- Digital PCR (dPCR):
- Digital PCR divides the PCR reaction into thousands of tiny partitions, each containing a single DNA molecule.
- This method provides absolute quantification of target DNA molecules and is highly precise.
- Digital PCR is an advanced technique that partitions the PCR reaction into thousands or even millions of individual reactions.
- Each partition contains a single molecule or a small number of target DNA sequences.
- By counting the number of positive partitions, researchers can determine the absolute quantity of the target DNA in the sample.
- dPCR offers extremely high precision and sensitivity, making it valuable in applications requiring accurate quantification.
- Multiplex PCR:
- Multiplex PCR amplifies multiple target sequences in a single reaction.
- It's useful for detecting several pathogens or genetic markers simultaneously.
- Multiplex PCR allows for the simultaneous amplification of multiple target DNA sequences in a single reaction.
- Different pairs of primers specific to distinct target sequences are included in the reaction mix.
- This technique is especially valuable for diagnosing
diseases caused by multiple pathogens, such as respiratory infections or
sexually transmitted diseases, as well as for analyzing multiple genetic
markers in a single experiment.
- Nested PCR:
- Nested PCR involves two sets of primers used in two consecutive PCR reactions.
- It is employed when the target DNA is present in low concentrations or when specificity is crucial.
- Nested PCR is a modification of the standard PCR technique.
- It involves two successive PCR reactions.
- In the first reaction, a pair of outer primers is used to amplify the target DNA region.
- Then, in the second reaction, a pair of inner primers is used to amplify a smaller region within the first PCR product.
- Nested PCR
increases specificity and reduces the risk of amplifying nonspecific products,
making it useful in applications where high specificity is required, such as in
the detection of specific pathogens or rare genetic mutations.
- Hot Start PCR:
- This technique minimizes non-specific amplification by using a modified DNA polymerase that is inactive at lower temperatures.
- The enzyme is activated when the reaction is heated.
- Hot Start PCR is designed to reduce non-specific amplification and primer-dimer formation.
- It involves using a modified DNA polymerase that remains inactive at lower temperatures, preventing the extension of primers at non-specific sites during the initial setup of the PCR reaction.
- Activation of the polymerase occurs only
after an initial denaturation step at a higher temperature, improving
specificity and the overall success of the reaction.
- In situ PCR:
- In situ PCR amplifies DNA within cells or tissues, allowing researchers to study the location and distribution of specific DNA sequences within biological samples.
- In Situ PCR is a technique used to amplify specific DNA sequences within cells or tissues while preserving their original location.
- This method is invaluable for studying the
spatial distribution of DNA sequences in biological samples and is commonly used
in research fields such as pathology, developmental biology, and forensics.
- Asymmetric PCR:
- Asymmetric PCR generates a large excess of one DNA strand over the other.
- It's often used in sequencing and cloning applications.
- Asymmetric PCR involves using an excess of one primer relative to the other.
- This results in
the preferential amplification of one DNA strand, which can be useful in
sequencing, cloning, or preparing single-stranded DNA for various applications.
- Long PCR:
- Long PCR is used to amplify longer DNA fragments, such as entire genes or genomes.
- Specialized DNA polymerases and reaction conditions are employed for this purpose.
- Long PCR is employed when amplifying long DNA fragments, such as entire genes or large genomic regions.
- Specialized DNA polymerases, optimized reaction conditions, and modified primers are used to facilitate the amplification of longer templates.
- Long PCR is essential in genome sequencing, cloning, and genetic analysis.
- High-Fidelity PCR:
- High-fidelity PCR employs DNA polymerases with proofreading capabilities to minimize the introduction of errors during DNA amplification.
- It's used when accuracy is crucial, such as in DNA sequencing or cloning experiments.
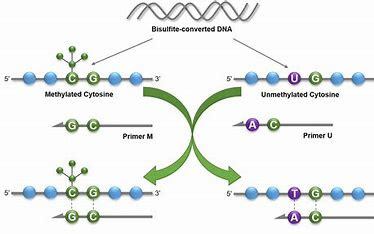
- Methylation-Specific PCR (MSP):
- MSP is designed to detect DNA methylation patterns, which can be associated with gene regulation and diseases like cancer.
- By using specific primers that target methylated or
unmethylated DNA regions, researchers can analyze the methylation status of specific
genes.
- Overlap Extension PCR (OE-PCR):
- Overlap Extension PCR is a technique used to create DNA sequences with specific overlaps between segments.
- It's often used in molecular cloning and site-directed mutagenesis to
join multiple DNA fragments together and create custom-designed genetic constructs.
- Other techniques and various fields
- Random Amplification of Polymorphic DNA (RAPD):
- RAPD is a PCR technique used for DNA fingerprinting and genetic diversity analysis.
- It
employs short, random primers to amplify genomic DNA, producing unique banding
patterns that can be used for genetic profiling and diversity assessment.
- PCR in Genetic Engineering:
- PCR plays a crucial role in genetic engineering by enabling the amplification of specific DNA fragments for subsequent manipulation and cloning.
- It is used to create recombinant DNA, perform site-directed mutagenesis, and construct genetically modified organisms (GMOs).
- Environmental DNA (eDNA) Analysis:
- PCR is used in eDNA analysis to detect the presence of specific organisms or pathogens in environmental samples, such as water or soil.
- It's a valuable tool for monitoring biodiversity and studying the distribution of species in ecosystems.
- Clinical Diagnostics and Pathogen Detection:
- PCR is widely used in clinical diagnostics to detect various pathogens, including bacteria, viruses, and fungi.
- It's critical for identifying the causative agents of
infectious diseases and tracking disease outbreaks.
- Pharmacogenomics:
- PCR is employed in pharmacogenomics to study how genetic variations influence an individual's response to drugs.
- By analyzing specific genetic markers, healthcare providers can personalize drug treatments for patients to optimize efficacy and minimize adverse effects.
- Field-Portable PCR Devices:
- Advances in PCR
technology have led to the development of field-portable PCR devices, which are
used in remote or resource-limited settings for rapid disease diagnosis,
environmental monitoring, and forensic investigations.















Comments
Post a Comment